Does the operon concept apply to Eukaryotes? Describe a suggested mechanism for gene regulation in Eukaryotes.
The operon system of gene regulation has proved not be applicable in eukaryotes. Crossing-over & chromosome structural changes (such as translocations, inversions) disrupt clusters of genes that otherwise might become operons. In prokaryotes, metabolically related genes are lying close together whereas in eukaryotes metabolically related genes may be scattered throughout the genome. Moreover, in a eukaryotic cell, compartmentalization is there. The nucleus contains most of the genetic material. The compartmentalization has a consequence for gene expression. Regulatory processes in a eukaryotic act, or might act, to control gene expression at various levels. Various potential control points recognized in eukaryotes are regulation at the level of gene structure, initiation of transcription, processing transcript & transport to the cytoplasm, translation of mRNA & post-translation control.
Gene regulation in eukaryotes:
The regulation in higher eukaryotes can be either short-term or long-term. In higher eukaryotes, one of the well-studied systems of short-term gene regulation is the control of enzyme synthesis by steroid hormones. And the long-term regulation of gene expression refers to the regulation during the development & differentiation of organisms.
One of the most popular models of regulation of gene expression in eukaryotes is the Davidson-Britten model.
Britten-Davidson model:
R.J Britten & E.H Davidson developed a theoretical model in 1969 to explain enormous genome & possible modes of regulation in higher forms. This model was later elaborated by them in 1973. The model assumes the presence of the following four type of sequence: Producer gene, Receptor site (gene), Integrator gene & Sensor site.
Transcription of a producer gene could occur only if at least one of its receptor sites was activated by forming a sequence-specific complex with activator RNA (or activator protein). This RNA (or protein) would be synthesized by integrator gene in response to the signals by the sensor site that are sensitive to external or internal development signals.
In Britten-Davidson model, it is also proposed that receptor sites & integrator genes may be repeated a number of times so as to control the activity of a large number of genes in the same cell. Repetition of receptor ensures that same activator recognizes all of them & in this way several enzymes of one metabolic pathway are simultaneously synthesized. Transcription of the same gene may be needed in different developmental stages. This is achieved by the multiplicity of receptor sites & integrator genes. Each producer gene may be several receptor sites, each responding to one activator. Thus a single activator though can recognize several genes but different activators may activate the same gene at a different time. An integrator gene may also fall in a cluster with the same sensor site.
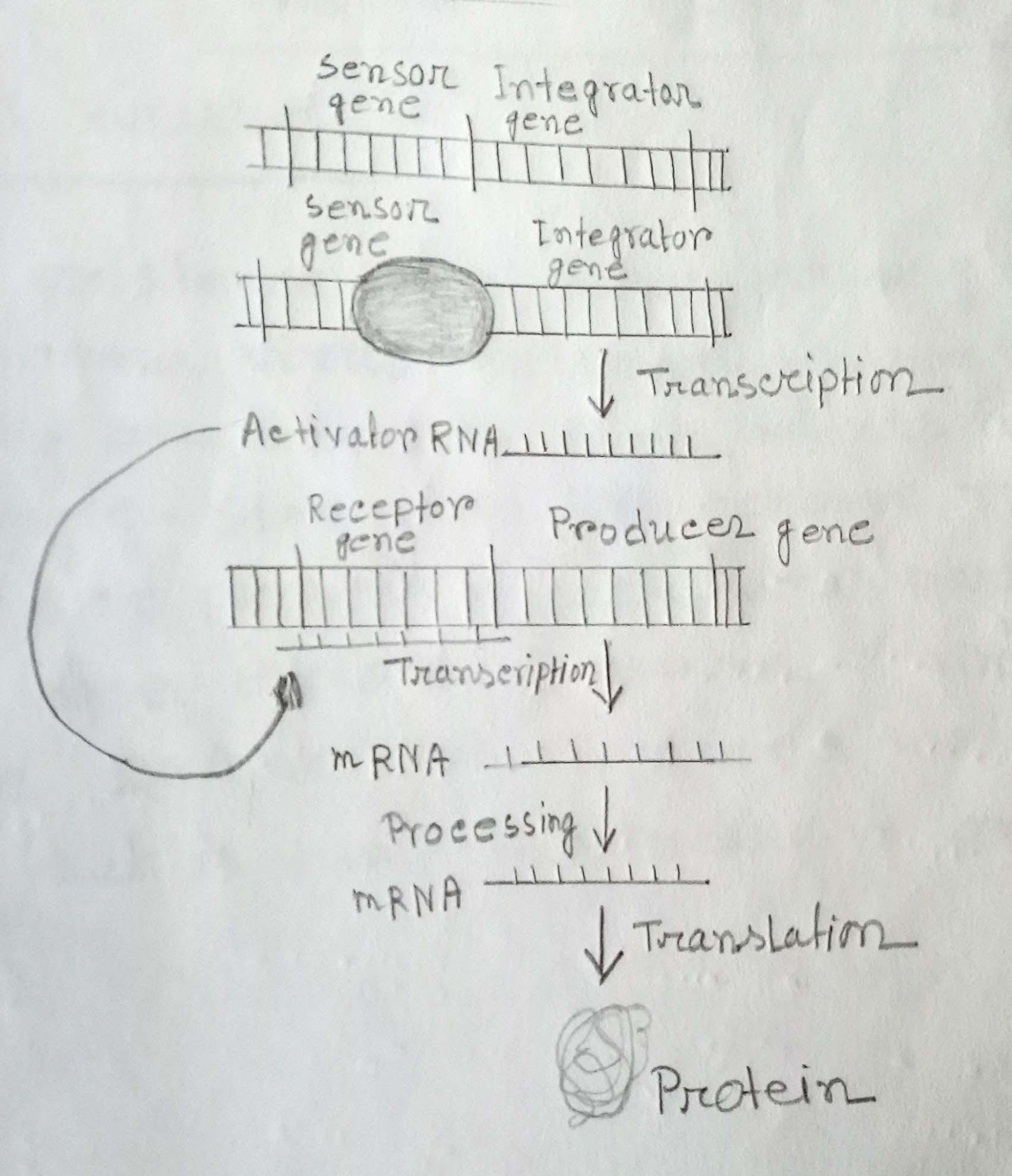
Fig: Britten-Davidson model of eukaryotic gene regulation. Interactions among different components of the machinery are shown.
Britten-Davidson model is also known as gene battery model. A set of structural genes controlled by one sensor site is termed as a battery. Sometimes when major changes are needed, it is necessary to activate several sets of genes. If one sensor site is associated with several integrators, it may be caused transcription of all integrators simultaneously thus causing transcriptions of several producer genes through receptor sites. The repetition of integrator genes & receptor sites is consistent with the reports that sufficient repeated DNA occurs in the eukaryotic cells.