Using diagrams explain the Holliday Model for the molecular mechanism of recombination
Holliday Model One model for eukaryotic DNA recombination that survives was proposed by Robin Holliday in 1964. This pathway begins when an endonuclease cleaves single strands of each of the two parental DNA molecules. Segments of the single strands on one side of each cut are displaced from their complementary strands. In this process, DNA-binding proteins, helix destabilization proteins, & DNA unwinding proteins or DNA helicases are also said to be involved.
This displaced strands then exchange pairing partners, base-pairing with the intact complementary strands of the homologous chromosomes. This process is also aided by certain proteins such as Rec A protein. The rec A protein mediates such a reaction by binding to the unpaired strand of DNA, searching for a homologous DNA sequence. A homologous double helix is found, promoting the displacement of a segment of one strand of the double helix by the unpaired strand.
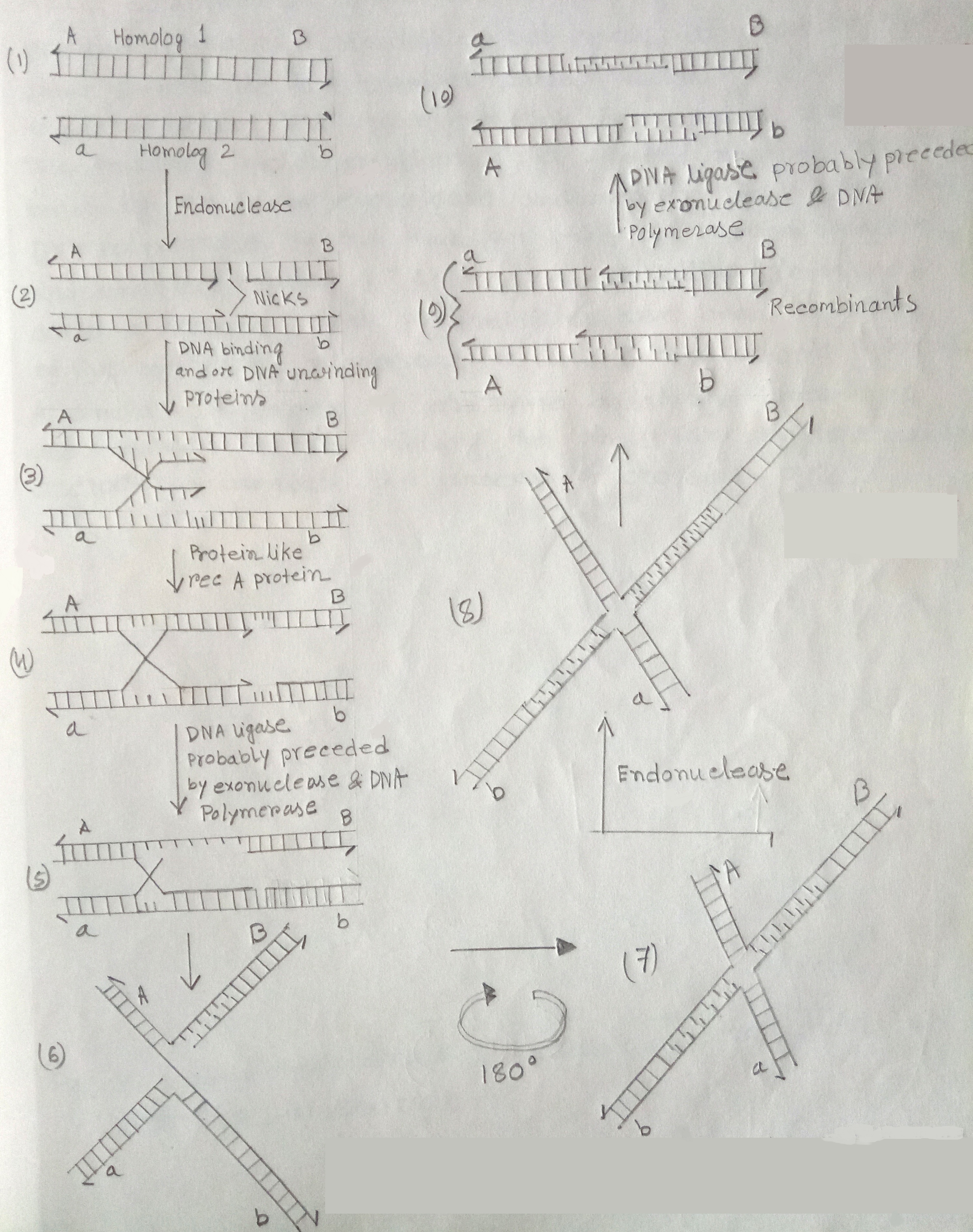
Fig: Pathway for the occurrence of crossing over by breakage & reunion based on the model of Robin Holliday
The cleaved strands are then covalently joined in recombinant arrangements by the enzyme DNA ligase. If the original breaks in the two strands do not occur at exactly the same site in the two homologs some, 'tailoring' will be required before DNA ligase catalyzes the reunion process. Thus tailoring involves excision of a limited number of bases by an exonuclease & repair synthesis by a DNA polymerase. By this time an x-shaped recombination intermediate called a 'chi' form or Holliday intermediate is formed. Such intermediated have been observed by the electron microscopy in several prokaryotic systems. A similar sequence of enzyme catalyzed breakage & reunion events involving the other two single strands occurs to complete the process of crossing-over.
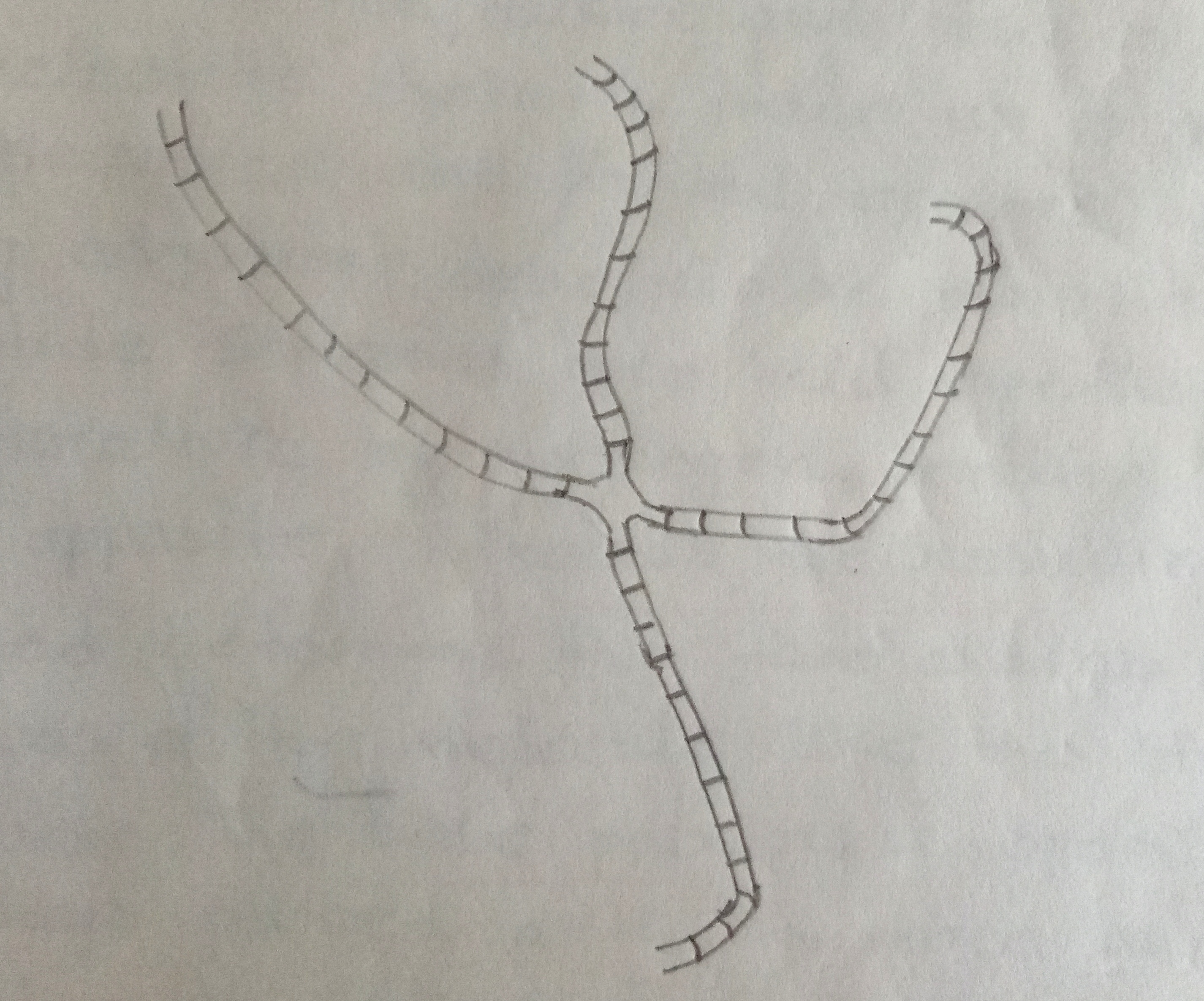
Fig: X-shaped recombinant intermediate as seen under the electron microscope.